Large insert bacterial artifical chromosome libraries (BACS) are invaluble tools and a source of genomic DNA for physical mapping, positional cloning and as a scaffold for whole genome sequencing. Rubus idaeus is an ideal candidate for BAC library construction, since it is diploid (2n=2x=14) and has a very small genome (275 Mbp). Indeed, the genome size of raspberry is only twice that of the model plant Arabidopsis, making it highly amenable to complete physical map construction, and thereby providing a platform for map-based gene cloning and comparative mapping with other members of the Rosaceae.
The isolation of high-molecular-weight DNA (HMW-DNA), either in the form of embedded protoplasts or nuclei, is one of the most challenging steps required for the construction of plant large insert genomic libraries. Whereas, numerous buffer systems and HMW-DNA preparation methods have been shown to be applicable to some plant species, biochemical and morphological diversity within the plant kingdom are obstacles to the development of one single universal protocol. Raspberry and other soft-fruit species have proven recalcitrant to standard genomic DNA extractions as they contain very high levels of carbohydrates, particularly polysaccharides, and polyphenolic compounds. They require heavily modified methods for ordinary genomic DNA isolations and the utilization of activated charcoal in tissue culture to prevent growth inhibition due to excess polyphenolics released into the medium. To prepare HMW-DNA suitable for the construction of BAC libraries we have developed a novel nuclei isolation procedure based on a combination of nylon filters and Percoll gradients to purify nuclei extracts prior to embedding in agarose plugs (Hein et al. 2005).
Raspberry nuclei isolated with this method showed few signs of oxidation and contained high quality HMW-DNA which has been used for the construction of the first publicly available red raspberry BAC library covering 6-7 times the genome equivalent. At present the library comprises over 15,000 clones with an average insert size of approximately 130 kb and less than 1% contamination with chloroplast and mitochondrial DNA respectively.
Ingo Hein & Sandie Williamson
Contact: Ingo Hein for further details
Some illustrations from this work

Figure 1. Nylon membrane (40 micron) before (left) and after (right) filtration of nuclei homogenate. The brown staining, typically observed after filtration of raspberry or blackcurrant nuclei suspension, most likely consists of withheld polysaccharide and polyphenolics, which are responsible for the colouration.

Figure 2. Typical example of embedded raspberry nuclei following extraction with standard protocols (left) and the method described here (right). The browning of the agarose plugs is caused by co-precipitated and oxidising polyphenolics, which covalently bind to proteins and DNA.
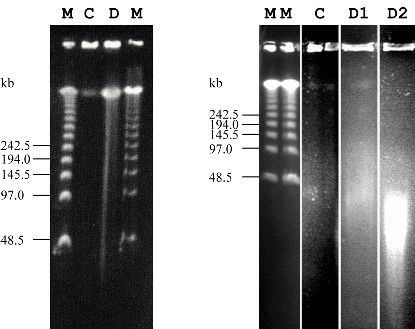
Figure 3. Restriction enzyme digests (HindIII) of raspberry HMW-DNA (left) and blackcurrant HMW-DNA (right). Lambda Ladder PFG marker (M) was run alongside undigested DNA (C) and digested HMW-DNA (D). The size of marker fragments is shown in kilobases (kb). Raspberry HMW-DNA was subjected to 0.5 - 80 U of HindIII and digested for 20 min at 37C. Blackcurrant HMW-DNA was digested with 20 - 120 U of HindIII and digested for 120 min (D1) and 24 h (D2) at 37C. The visualised digested DNA represents a mixture of different amounts of added enzyme
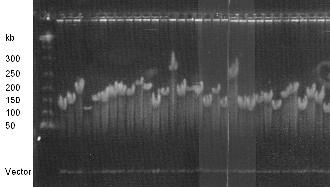
Figure 4. Sizing of 32 randomly selected BAC clones by NotI restriction enzyme digest. NotI restrictions sites are flanking the multi-cloning site of the vector pIndigoBAC-5 and release the cloned insert.
References
- Hein I, Williamson S, Russel J & Powell W (2005) Isolation of high molecular weight DNA suitable for BAC library construction from woody perennial soft-fruit species. BioTechniques 38:69-71.
